18 Term Enrichment Plots
Outside the scope of GO and KEGG ontologies, there are many other databases for which we can also compute functional enrichment for. This can be easily done using the enrichR package, which is an R interface to their website, Enrichr. This package allows to query a list of genes to more than 100 databases and retrieve the terms that are enriched for the list of genes, together with an adjusted p-value. The output of this process can be used with SCpubr::do_TermEnrichmentPlot() for its visualization.
Here is an example of how to run Enrichr to get the object that we need for SCpubr::do_TermEnrichmentPlot().
# Set necessary enrichR global options. This is copied from EnrichR code to avoid having to load the package.
suppressMessages({
options(enrichR.base.address = "https://maayanlab.cloud/Enrichr/")
options(enrichR.live = TRUE)
options(modEnrichR.use = TRUE)
options(enrichR.sites.base.address = "https://maayanlab.cloud/")
options(enrichR.sites = c("Enrichr", "FlyEnrichr", "WormEnrichr", "YeastEnrichr", "FishEnrichr"))
# Set the search to Human genes.
enrichR::setEnrichrSite(site = "Enrichr")
websiteLive <- TRUE
dbs <- enrichR::listEnrichrDbs()
# Get all the possible databases to query.
dbs <- sort(dbs$libraryName)
})
# Choose the dataset to query against.
dbs_use <- c("GO_Biological_Process_2021",
"GO_Cellular_Component_2021",
"Azimuth_Cell_Types_2021")
# List of genes to use as input.
genes <- c("ABCB1", "ABCG2", "AHR", "AKT1", "AR")
# Retrieve the enriched terms.
enriched_terms <- enrichR::enrichr(genes, dbs_use)For this chapter, we have generated the output for the previous code chunk. By default, SCpubr::do_TermEnrichmentPlot() returns a list with as many plots as databases were queried in Enrichr, or a single plot if only was used.
Any of the previous option will return a list of plots. The plots can, then, be assembled together.
# Default plot.
p <- SCpubr::do_TermEnrichmentPlot(enriched_terms = enriched_terms)
p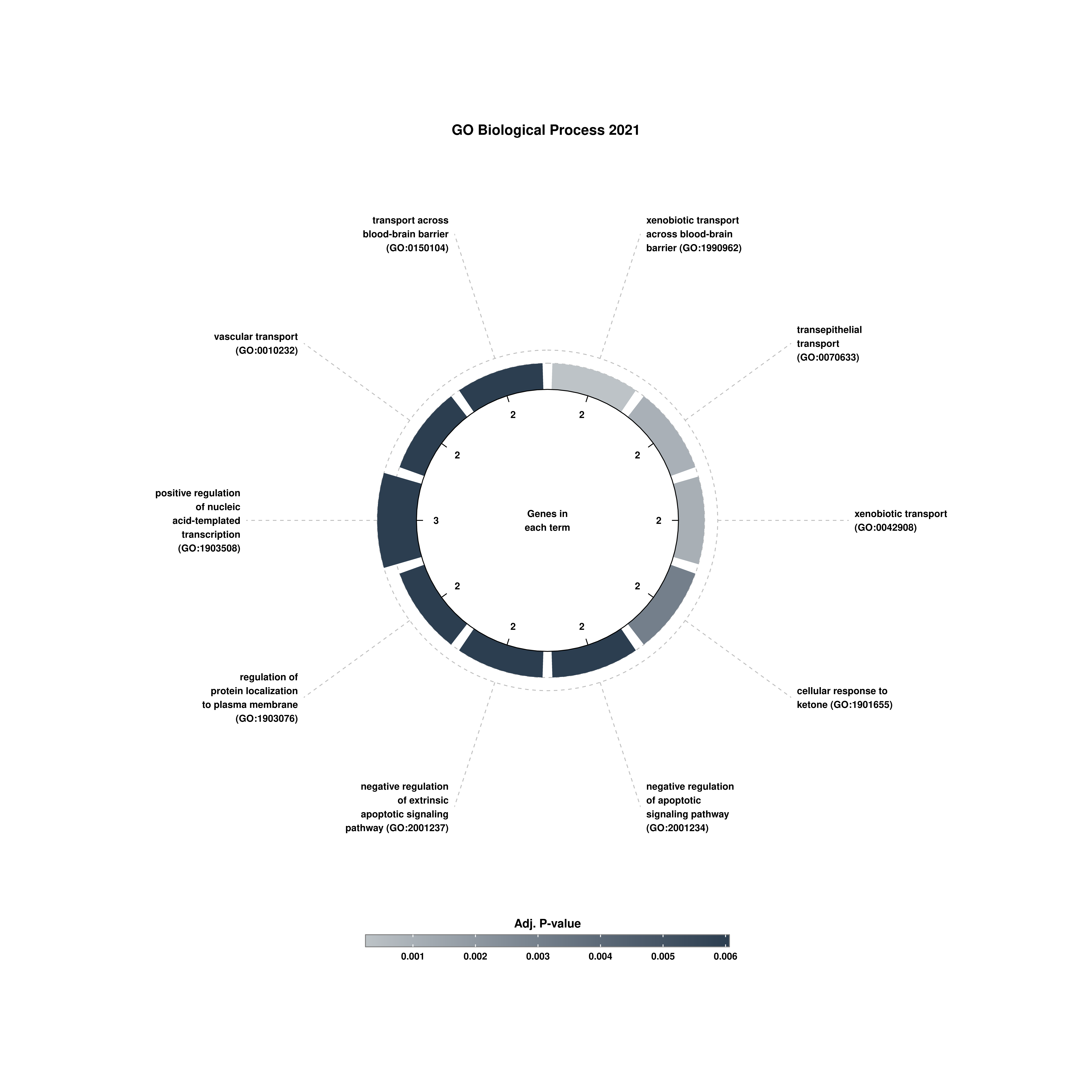
18.1 Modifying the number of terms to retrieve.
Depending on the focus of the analysis, we might want to only focus on one database but retrieve more terms from it. This can be achieved by using nterms parameter.
# Increased number of terms.
p <- SCpubr::do_TermEnrichmentPlot(enriched_terms = enriched_terms,
nterms = 15)
p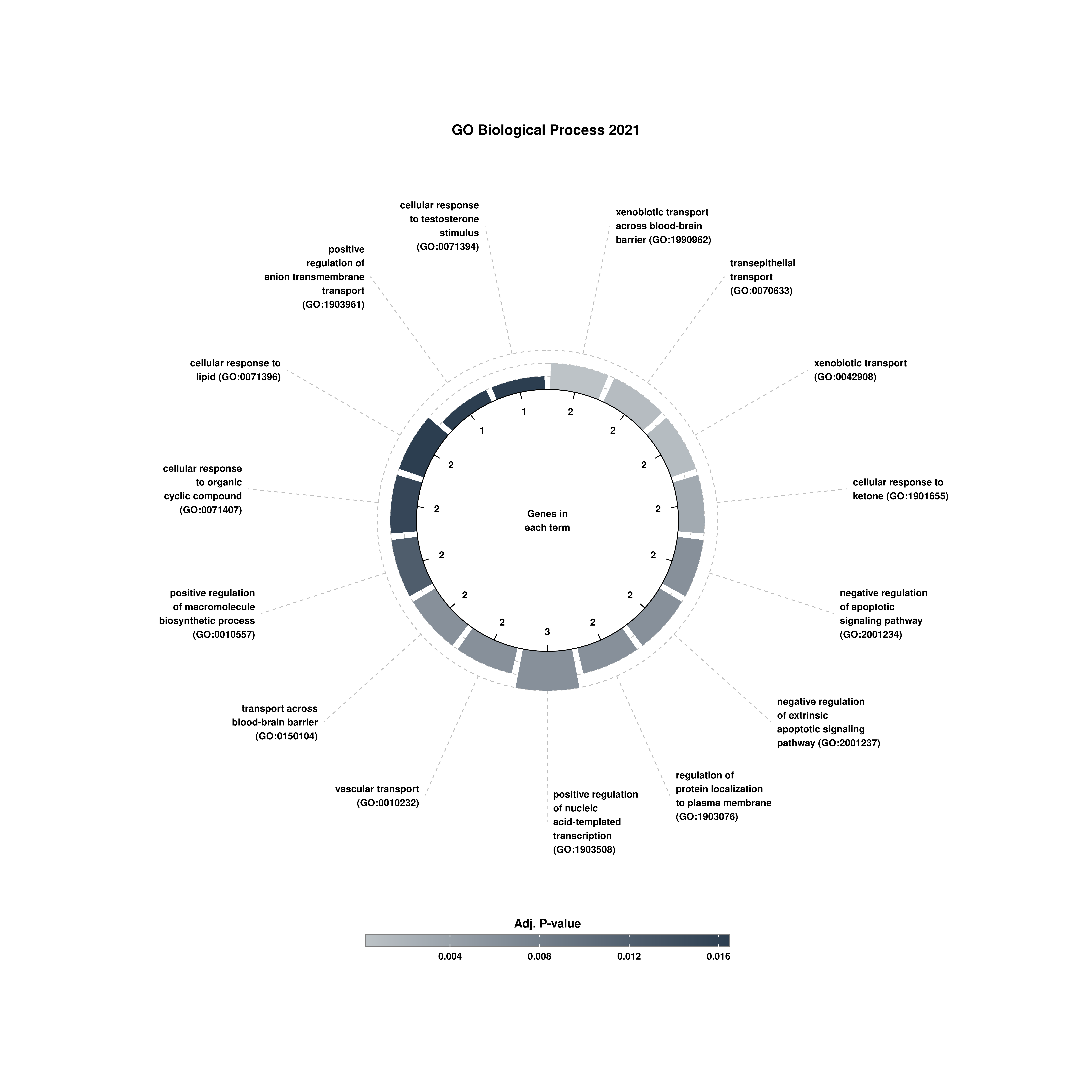
18.2 Modifying the length of the terms
Another issue with these plots is that, normally, the term itself takes too much space. For this, terms are wrapped according to a cutoff defined in nchar_wrap parameter. If the term has more characters than the value provided, it will be split more or less in half, always preserving whole words.
# Control the length of the terms.
p1 <- SCpubr::do_TermEnrichmentPlot(enriched_terms = enriched_terms,
nterms = 15)
p2 <- SCpubr::do_TermEnrichmentPlot(enriched_terms = enriched_terms,
nterms = 15,
nchar_wrap = 30)
p <- p1 / p2
p
In the same way, one can further enhance the limit in order to have each term in just one row.
18.3 Increase the font size of the labels
If you want to increase the font size of the labels - this is, anything that is not part of the legend or the titles, use text_labels_size parameter:
# Modify font size of the terms.
p1 <- SCpubr::do_TermEnrichmentPlot(enriched_terms = enriched_terms)
p2 <- SCpubr::do_TermEnrichmentPlot(enriched_terms = enriched_terms,
text_labels_size = 6)
p <- p1 / p2
p