6 Ridge plots
Ridge plots are a very interesting way to display multiple violin plots together. The main idea of this type of visualization is to observe shifts in the distribution of a given variable across several groups. This can be accomplished using package.
6.1 Basic usage
The most basic ridge plot can be computed as:
# Compute the most basic ridge plot.
p <- SCpubr::do_RidgePlot(sample = sample,
feature = "nFeature_RNA")
p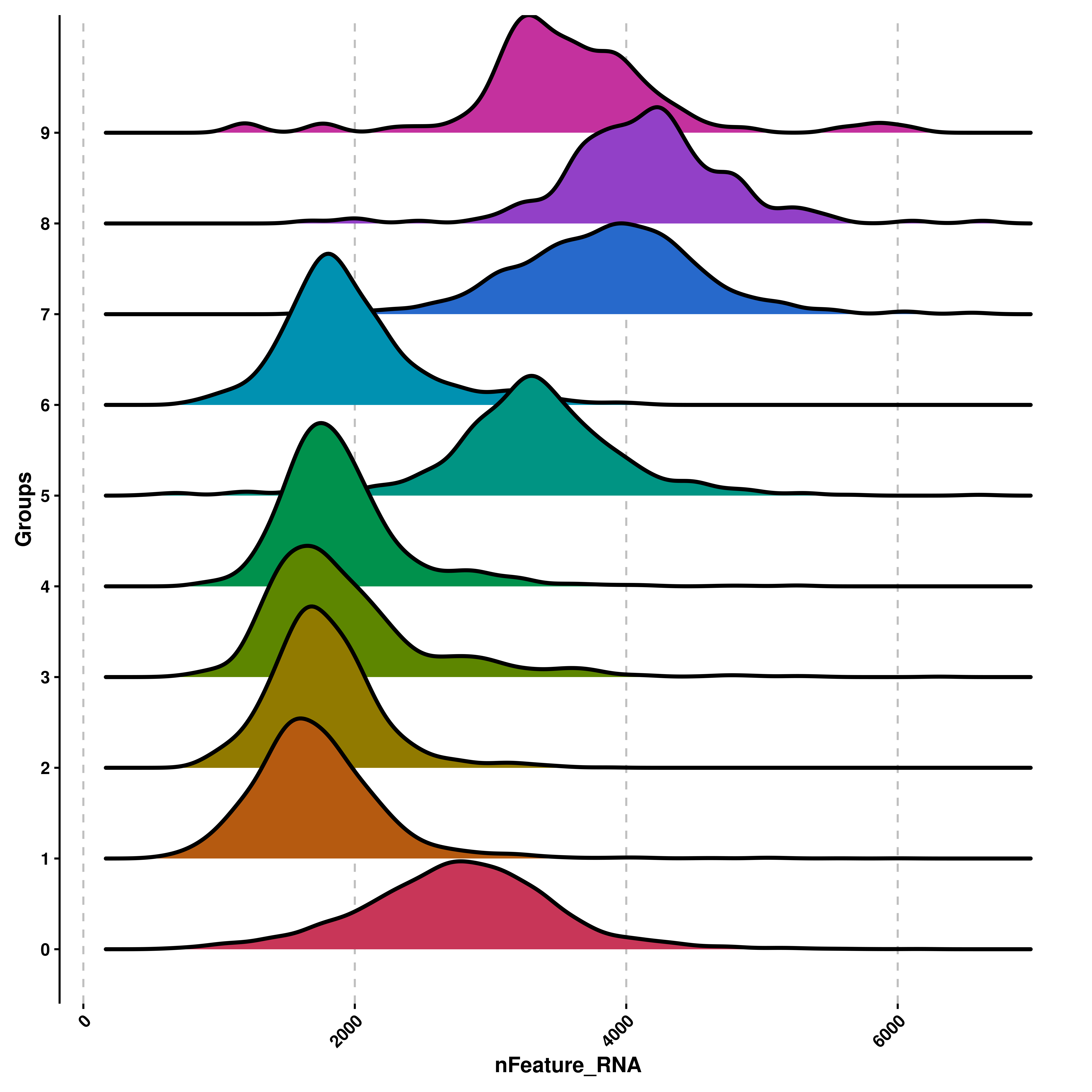
6.2 Use continuous color scales
By default, the groups plotted are the current identities in the sample, but this can be changed by using group.by. One can also color the ridges based on the continuous values being represented, using continuous_scale = TRUE. The direction of the color scale can be controlled using viridis_direction :
# Use continuous color scale.
p1 <- SCpubr::do_RidgePlot(sample = sample,
feature = "nFeature_RNA",
continuous_scale = TRUE,
viridis_direction = 1)
p2 <- SCpubr::do_RidgePlot(sample = sample,
feature = "nFeature_RNA",
continuous_scale = TRUE,
viridis_direction = -1)
p <- p1 / p2
p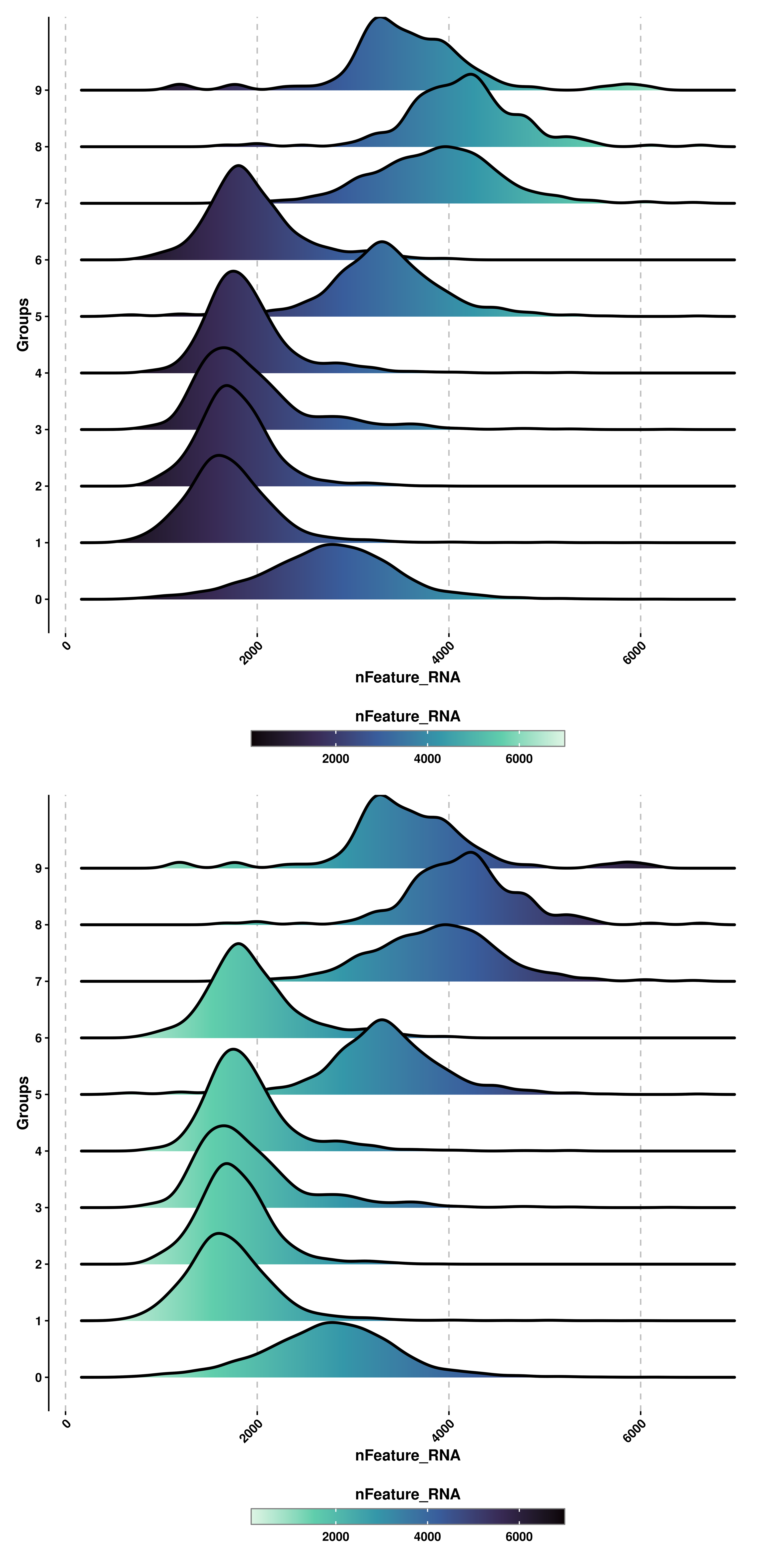
6.3 Plot quantiles of the distribution
One can also draw quantiles of the distribution for each of the groups. For this, we need to set up the scale to continuous. Also, we can modify the quantiles by using quantiles parameter:
# Draw quantiles of the distribution.
p1 <- SCpubr::do_RidgePlot(sample = sample,
feature = "nFeature_RNA",
continuous_scale = TRUE,
compute_quantiles = TRUE,
compute_custom_quantiles = TRUE)
p2 <- SCpubr::do_RidgePlot(sample = sample,
feature = "nFeature_RNA",
continuous_scale = TRUE,
compute_quantiles = TRUE,
compute_custom_quantiles = TRUE,
quantiles = c(0.1, 0.5, 0.75))
p <- p1 / p2
p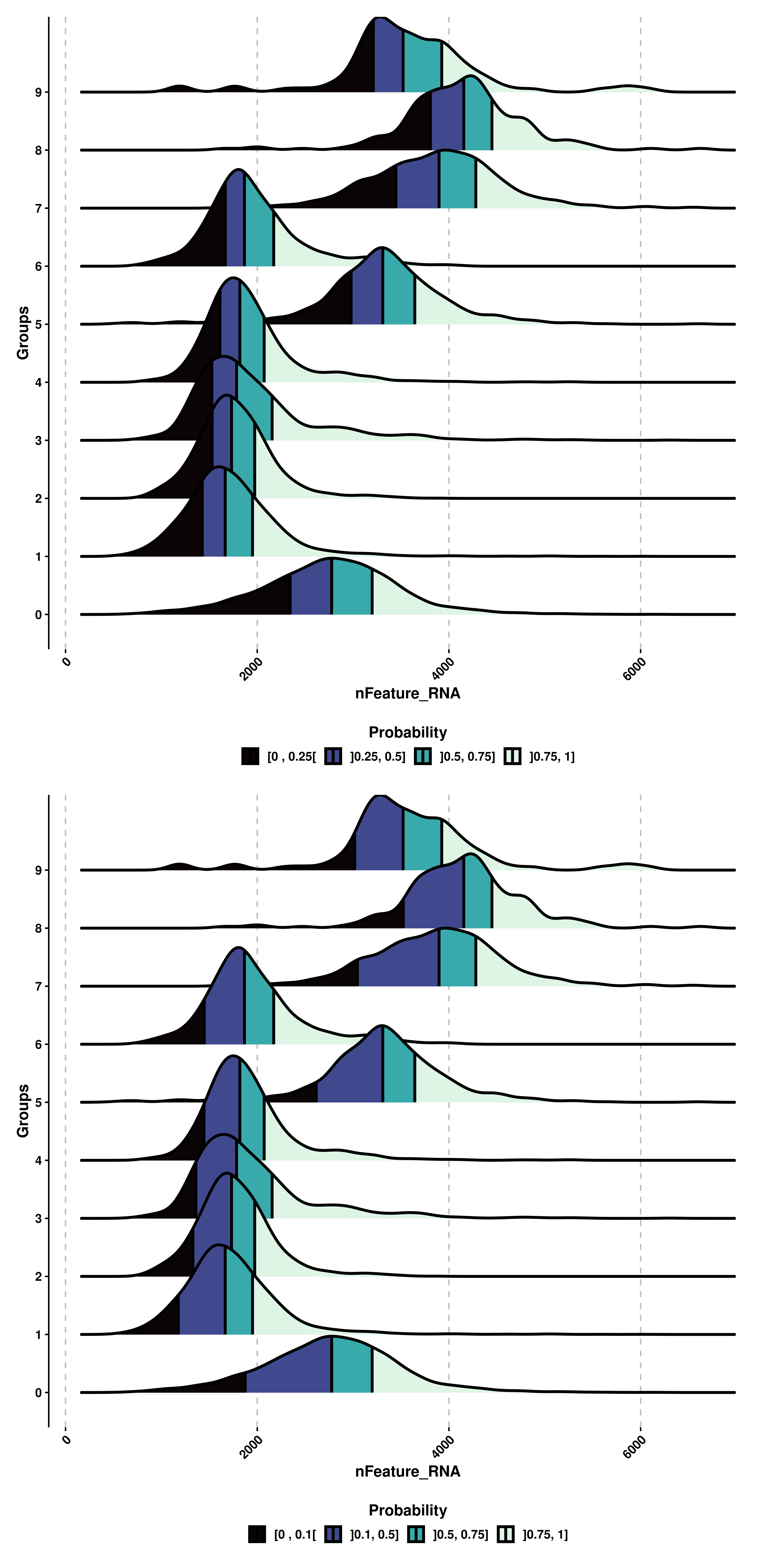
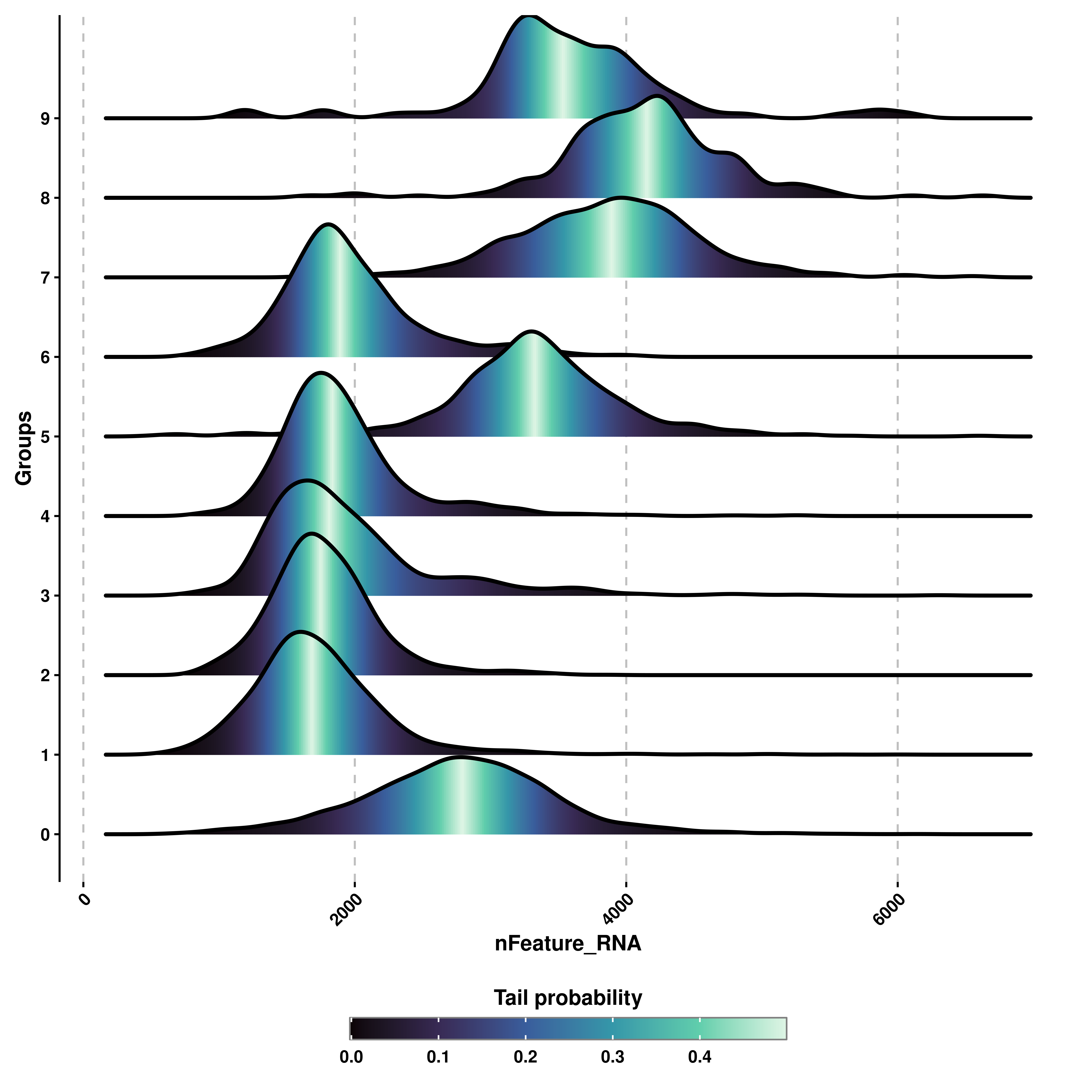
6.4 Compute probability tails
One can draw probability tails of the distribution by using compute_distribution_tails and prob_tails.
# Draw probability tails.
p1 <- SCpubr::do_RidgePlot(sample = sample,
feature = "nFeature_RNA",
continuous_scale = TRUE,
compute_quantiles = TRUE,
compute_distribution_tails = TRUE)
p2 <- SCpubr::do_RidgePlot(sample = sample,
feature = "nFeature_RNA",
continuous_scale = TRUE,
compute_quantiles = TRUE,
compute_distribution_tails = TRUE,
prob_tails = 0.3)
p <- p1 / p2
p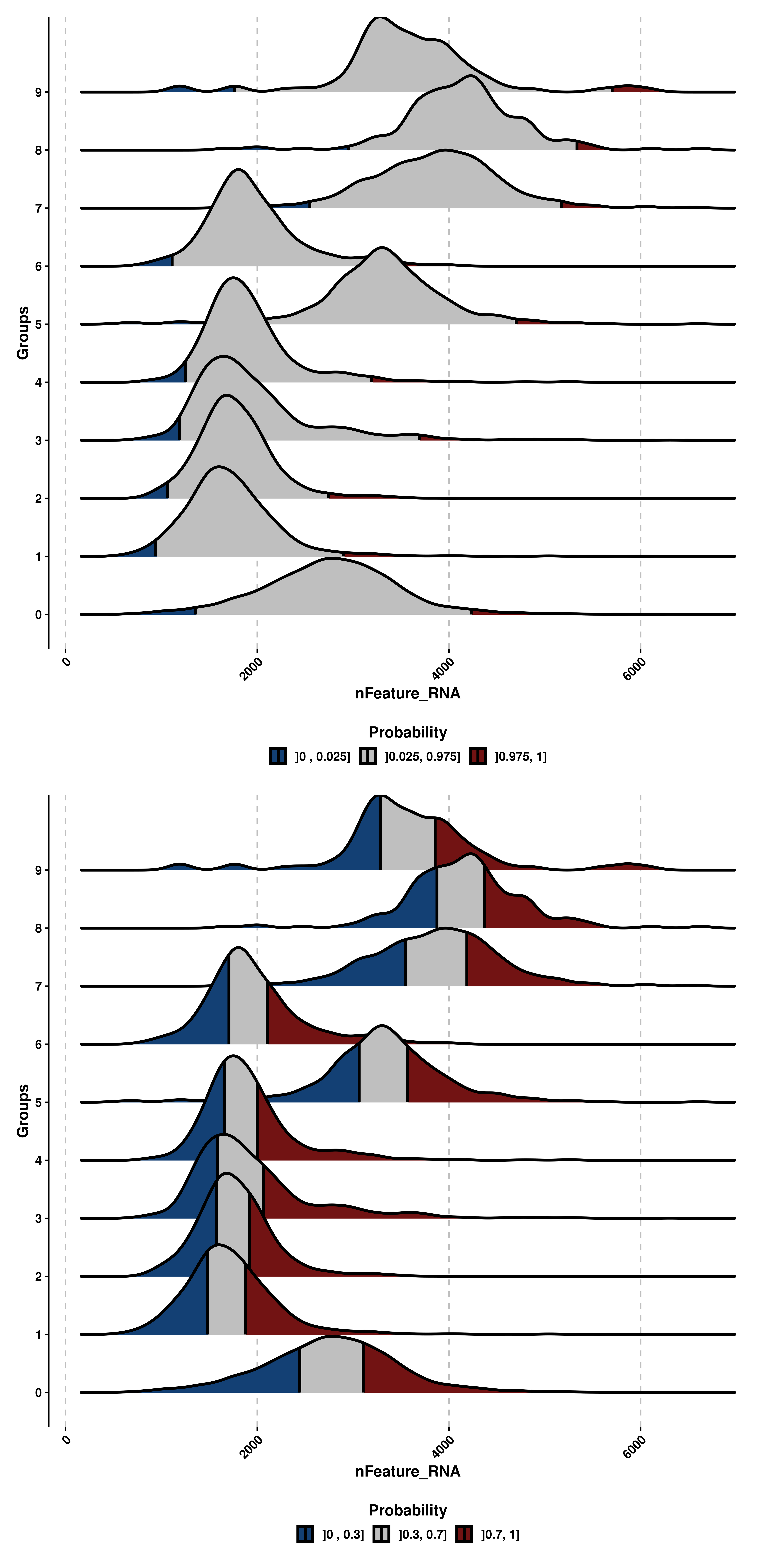
6.5 Compute probability densities
One can also display the probability densities of the distributions by using :
# Draw probability tails.
p <- SCpubr::do_RidgePlot(sample = sample,
feature = "nFeature_RNA",
continuous_scale = TRUE,
compute_quantiles = TRUE,
color_by_probabilities = TRUE)
p