SCpubr::do_FeaturePlot(sample = sample,
features = "nCount_RNA")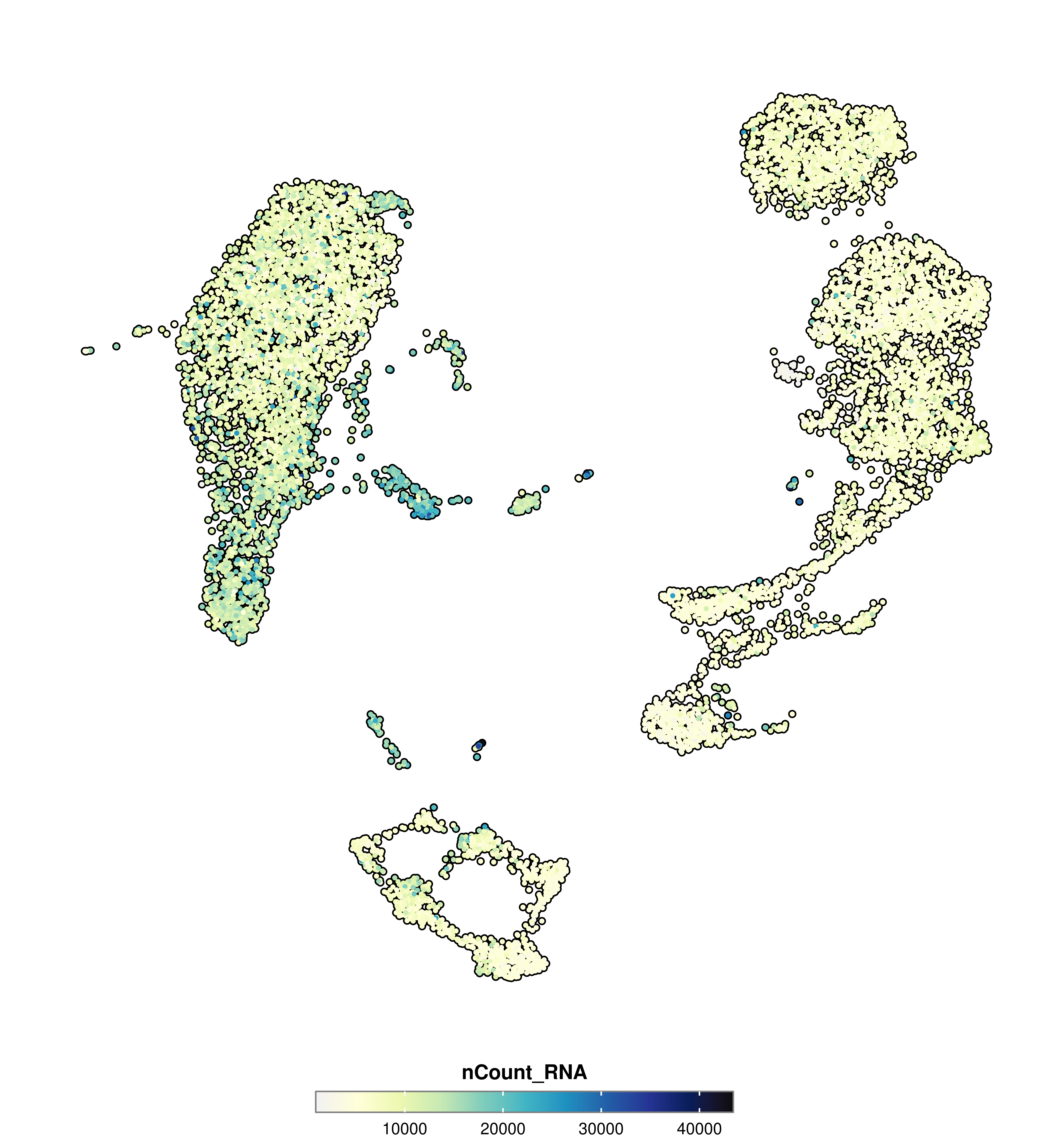
SCpubr::do_FeaturePlot(sample = sample,
features = "nCount_RNA")
SCpubr::do_FeaturePlot(sample = sample,
features = c("nCount_RNA", "nFeature_RNA"))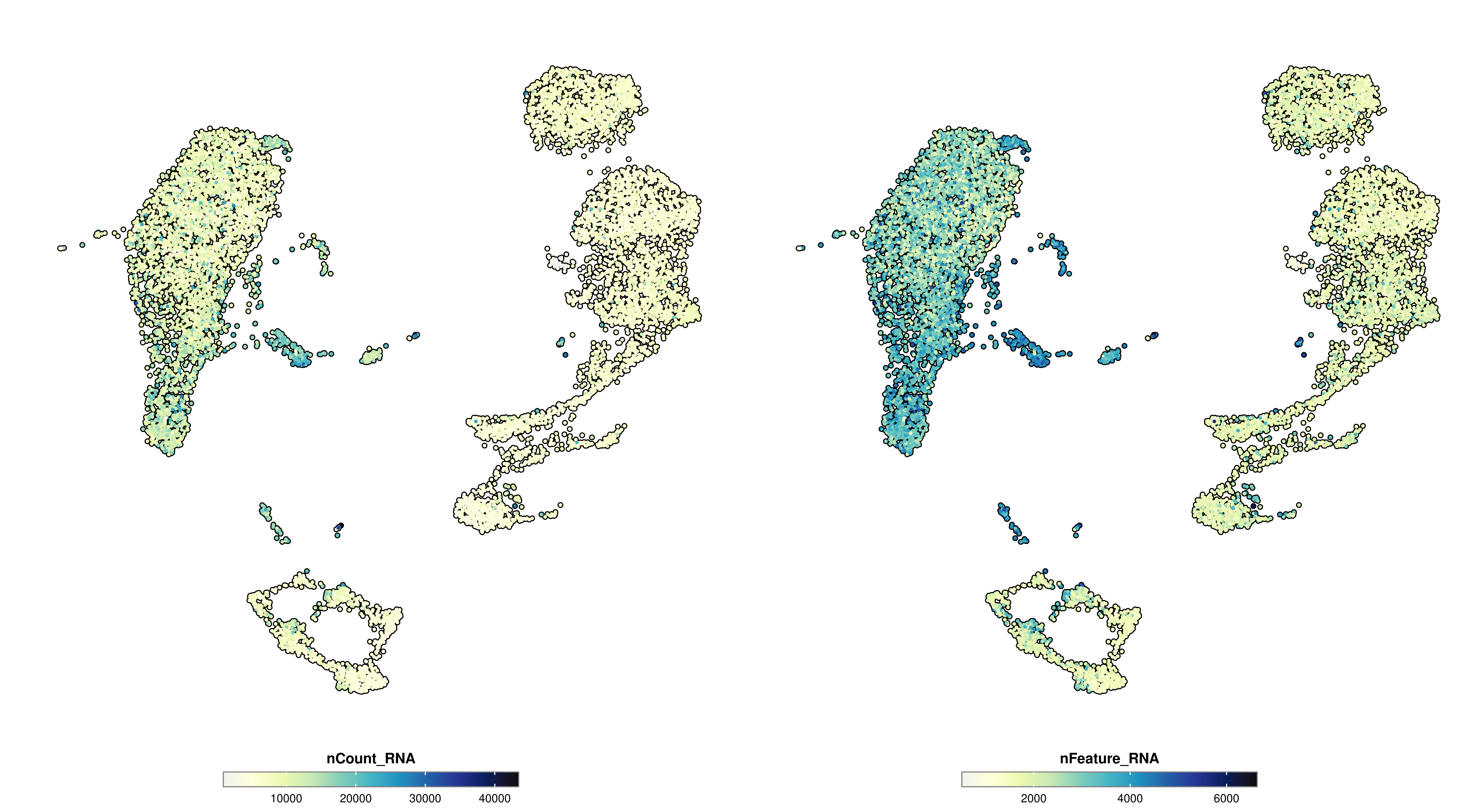
SCpubr::do_FeaturePlot(sample = sample,
features = "nCount_RNA",
min.cutoff = 5000,
max.cutoff = 30000)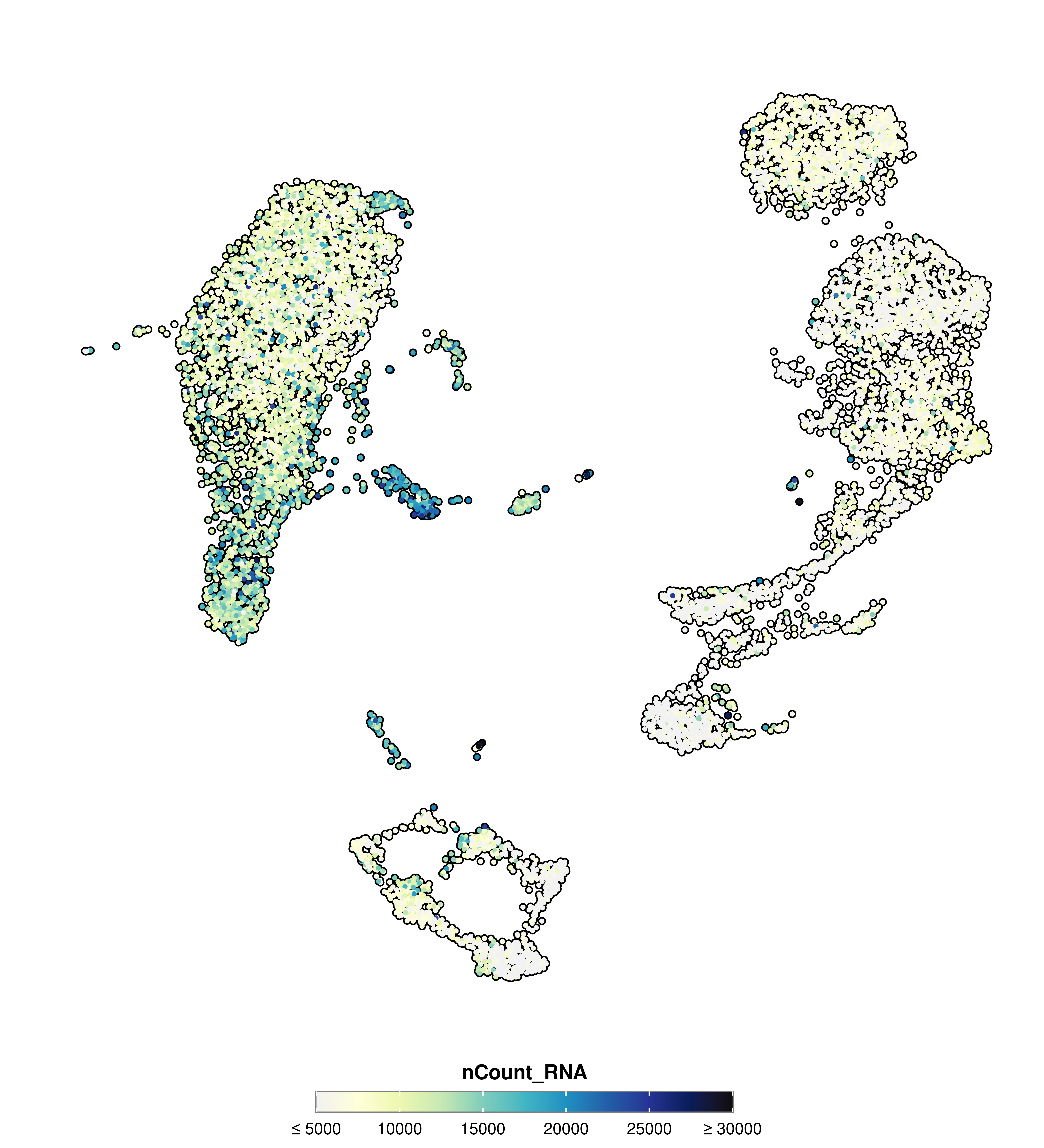
# Use a vector of cells
p1 <- SCpubr::do_FeaturePlot(sample = sample,
features = c("PC_1"),
cells.highlight = sample(colnames(sample), 500))
# Select identities.
p2 <- SCpubr::do_FeaturePlot(sample = sample,
features = c("PC_1"),
idents.highlight = c("0", "2", "5"))
p1 | p2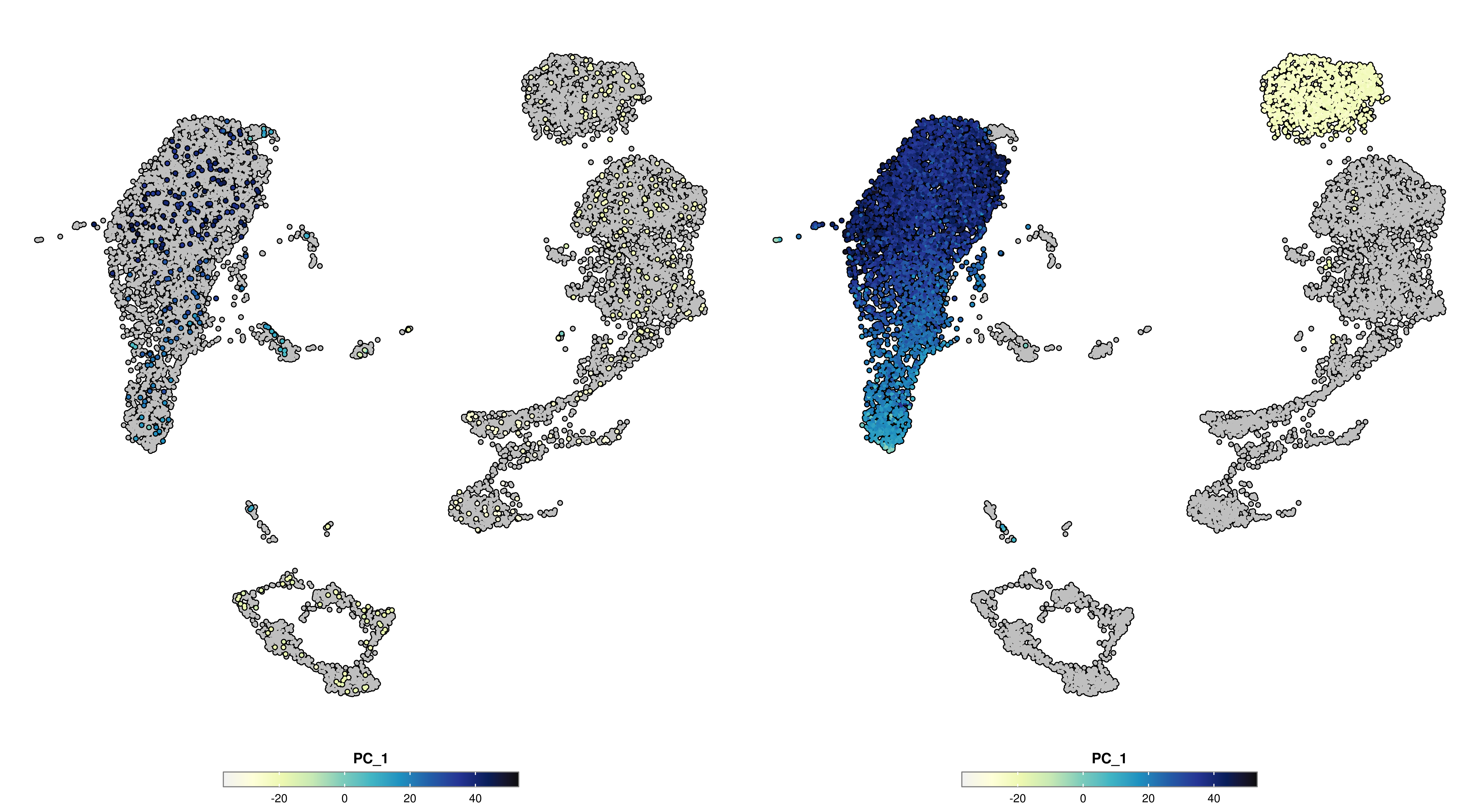
# Use a vector of cells
SCpubr::do_FeaturePlot(sample = sample,
features = c("PC_1"),
split.by = "seurat_clusters",
ncol = 5)
# Use a vector of cells
SCpubr::do_FeaturePlot(sample = sample,
features = c("PC_1"),
split.by = "seurat_clusters",
idents.keep = c("0", "5", "7"),
ncol = 3)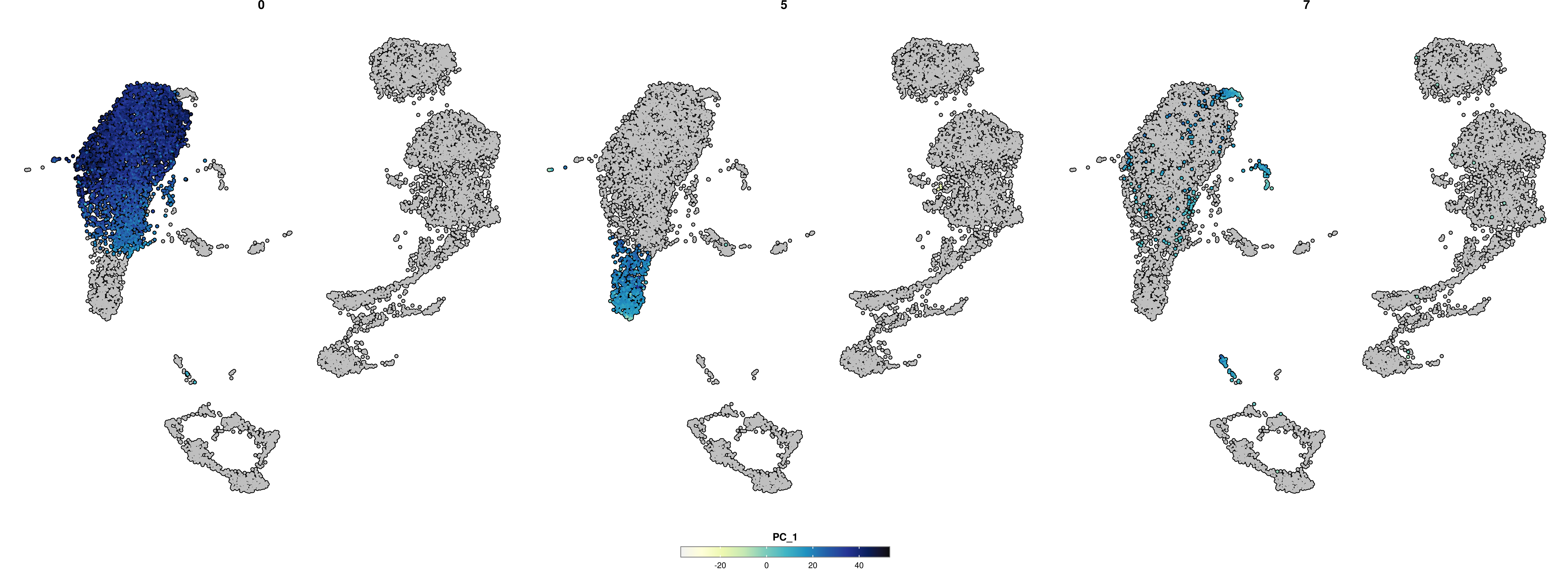
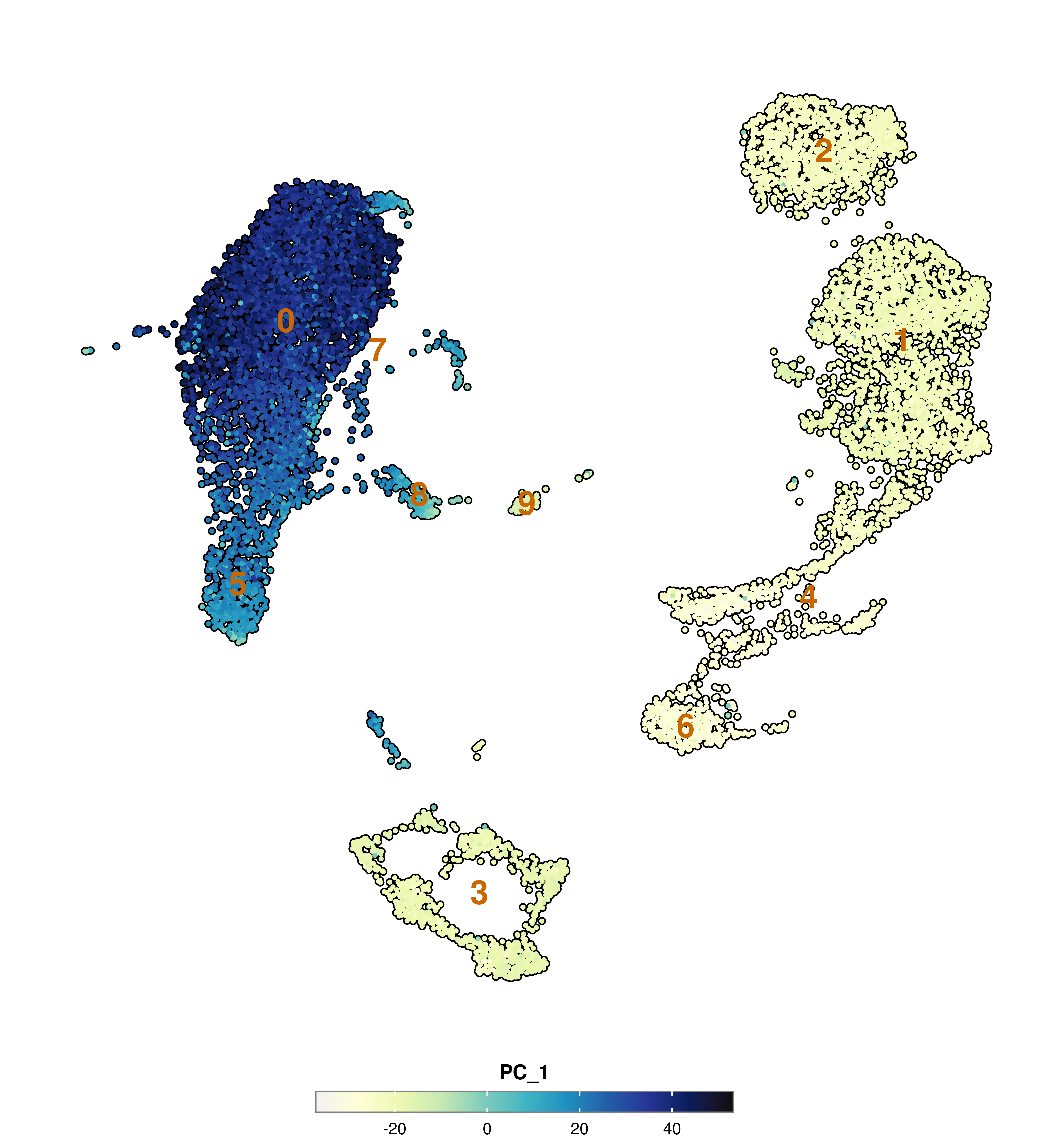
SCpubr::do_FeaturePlot(sample = sample,
features = "PC_1",
group.by = "seurat_clusters")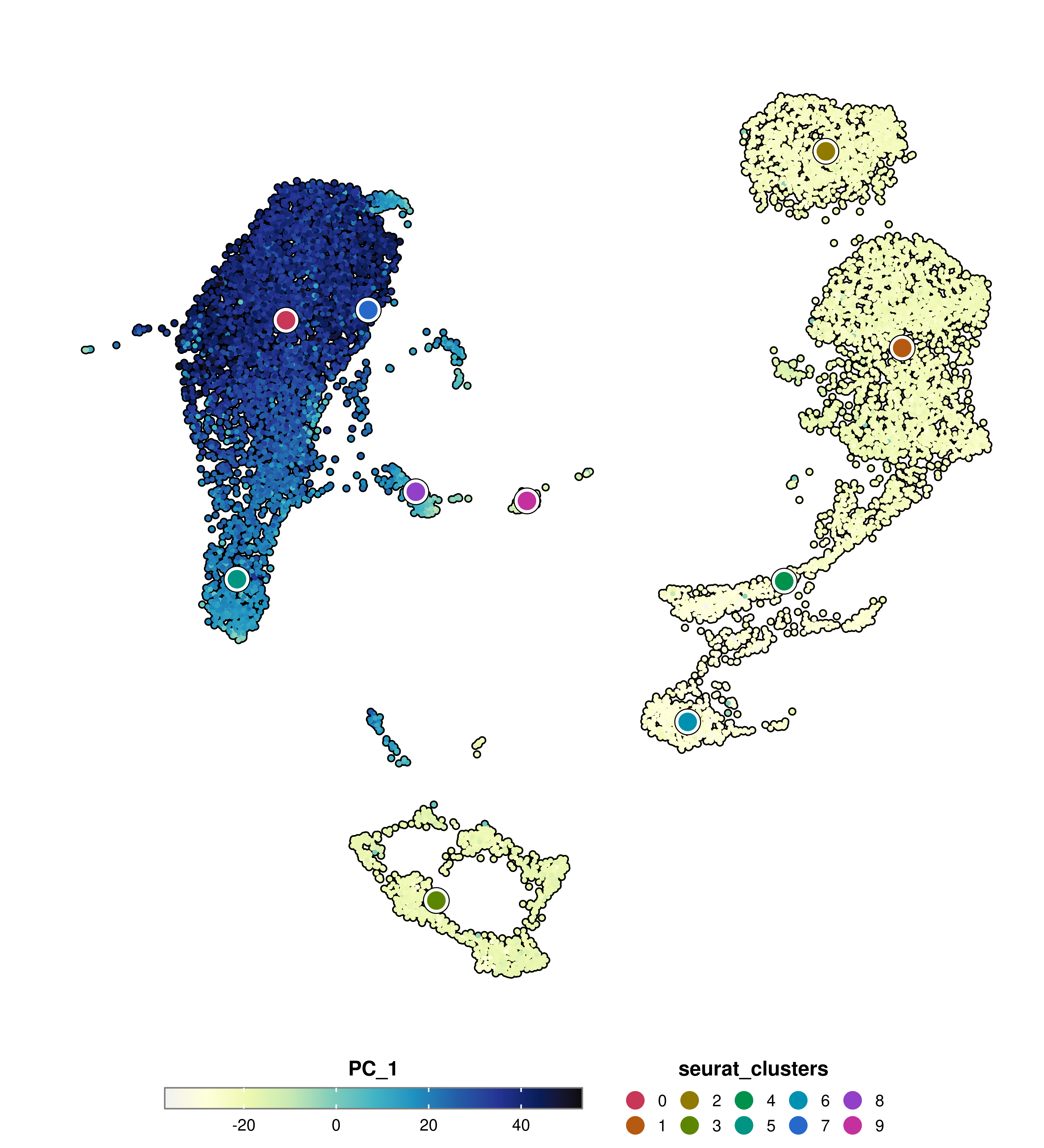
SCpubr::do_FeaturePlot(sample = sample,
features = "PC_1",
group.by = "seurat_clusters",
group.by.dot.size = 12)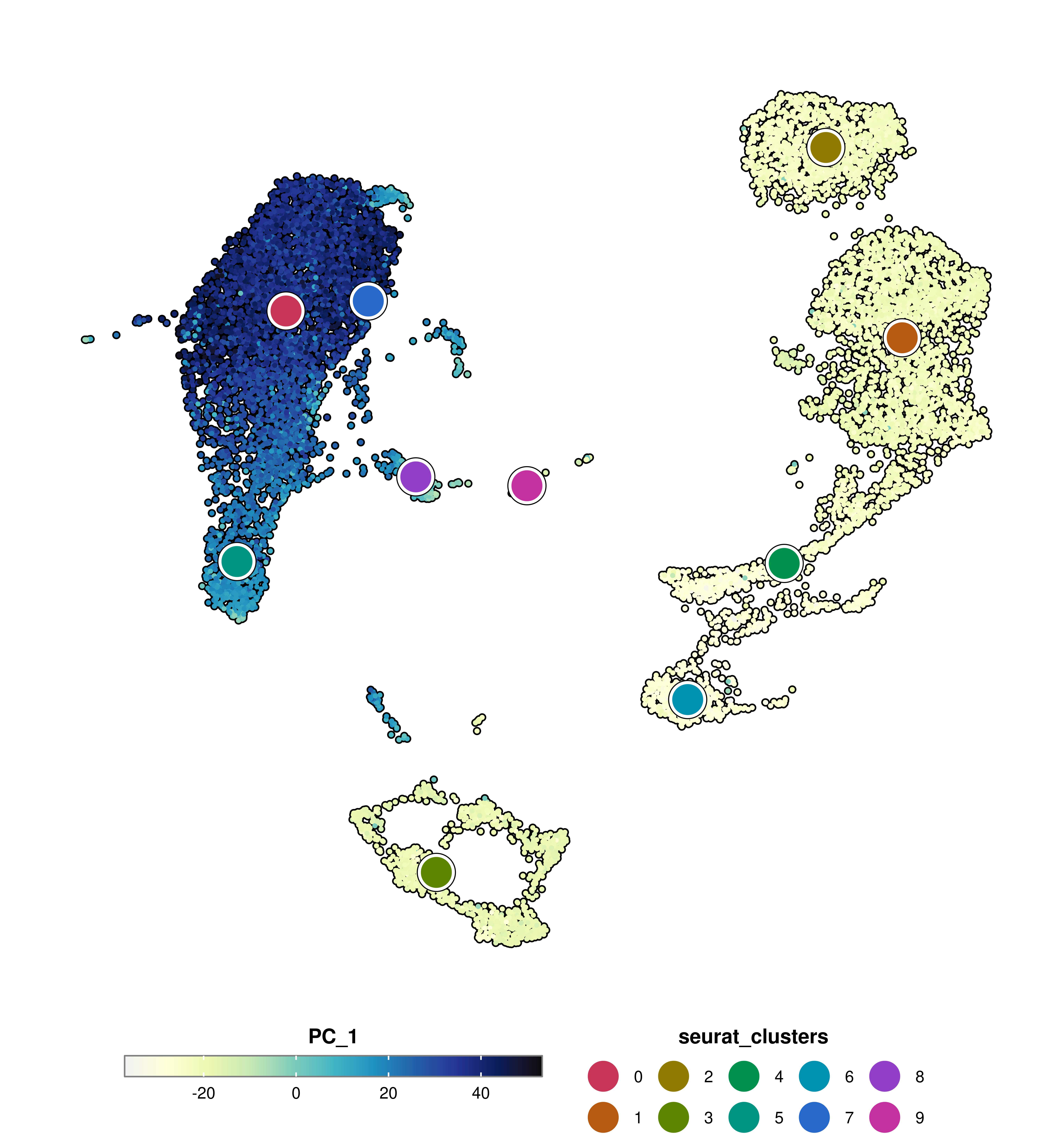
SCpubr::do_FeaturePlot(sample = sample,
features = "PC_1",
group.by = "seurat_clusters",
group.by.cell_borders = TRUE,
group.by.cell_borders.alpha = 0.1)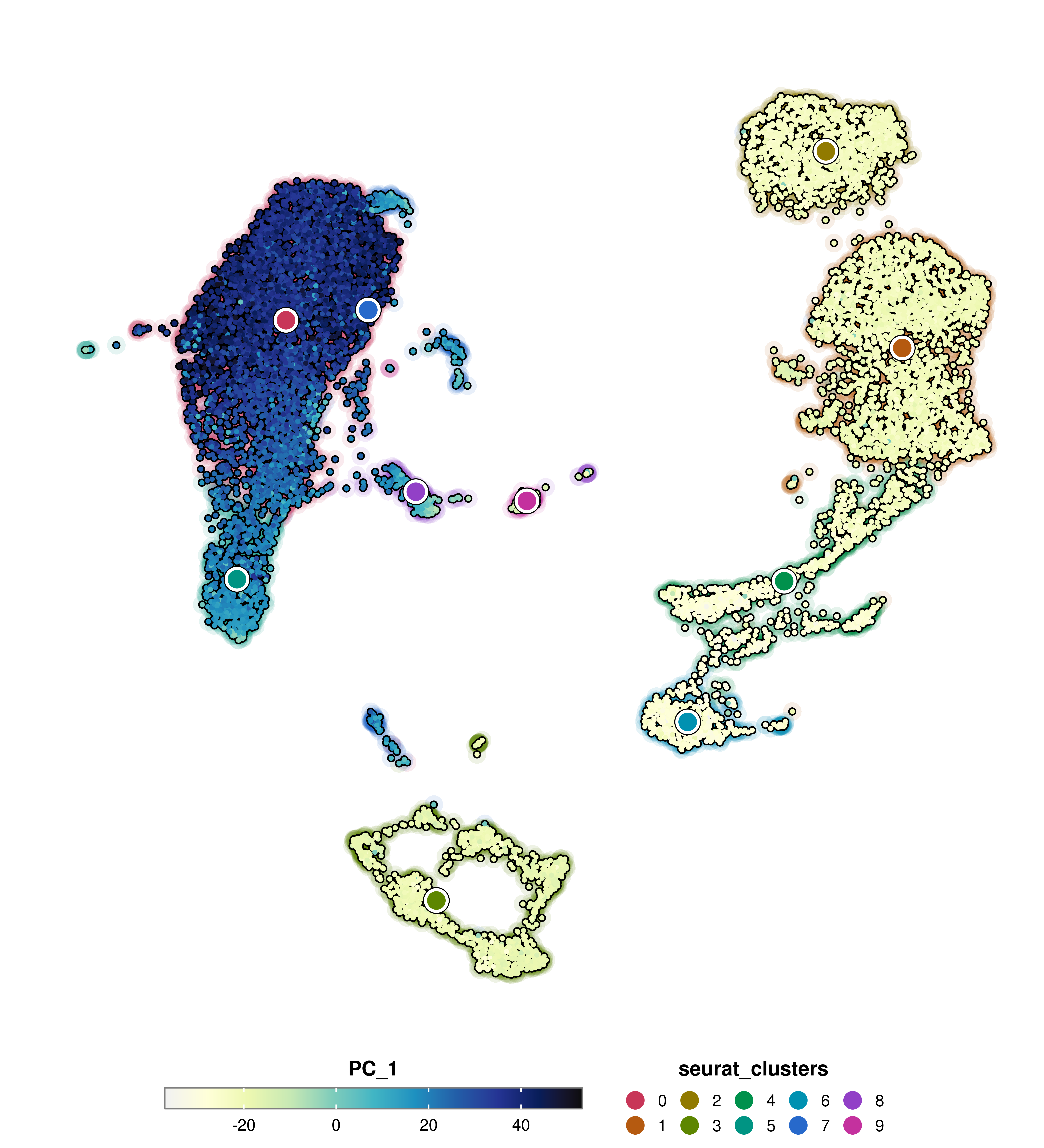
SCpubr::do_FeaturePlot(sample = sample,
features = "PC_1",
group.by = "seurat_clusters",
group.by.cell_borders = TRUE,
group.by.cell_borders.alpha = 0.1,
group.by.show.dots = FALSE)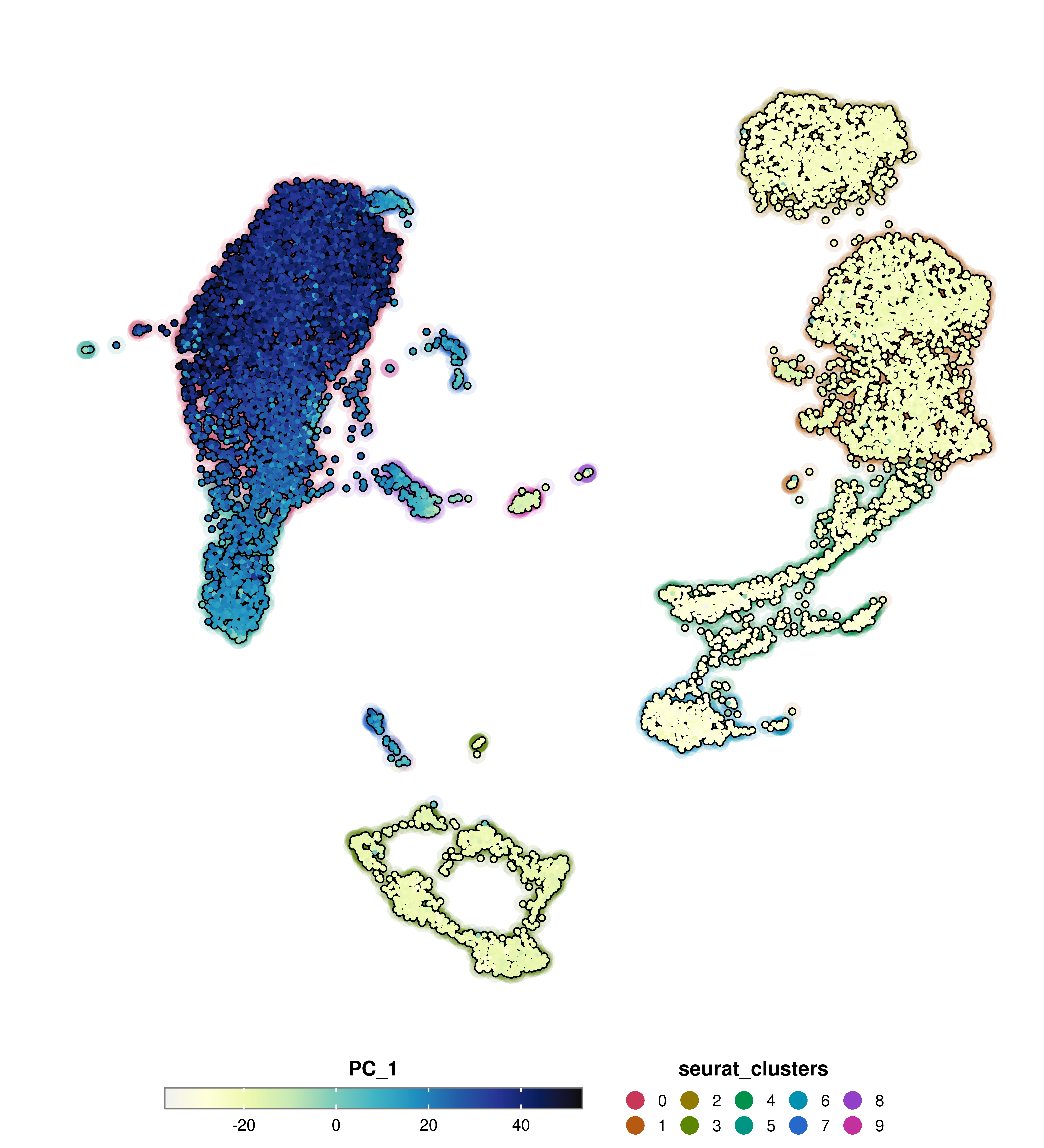
SCpubr::do_FeaturePlot(sample = sample,
features = "PC_1",
group.by = "seurat_clusters",
group.by.cell_borders = TRUE,
group.by.cell_borders.alpha = 0.1,
group.by.show.dots = TRUE,
group.by.legend = "My Clusters",
group.by.colors.use = c("0" = "#001219",
"1" = "#005f73",
"2" = "#0a9396",
"3" = "#94d2bd",
"4" = "#e9d8a6",
"5" = "#ee9b00",
"6" = "#ca6702",
"7" = "#bb3e03",
"8" = "#ae2012",
"9" = "#9b2226"))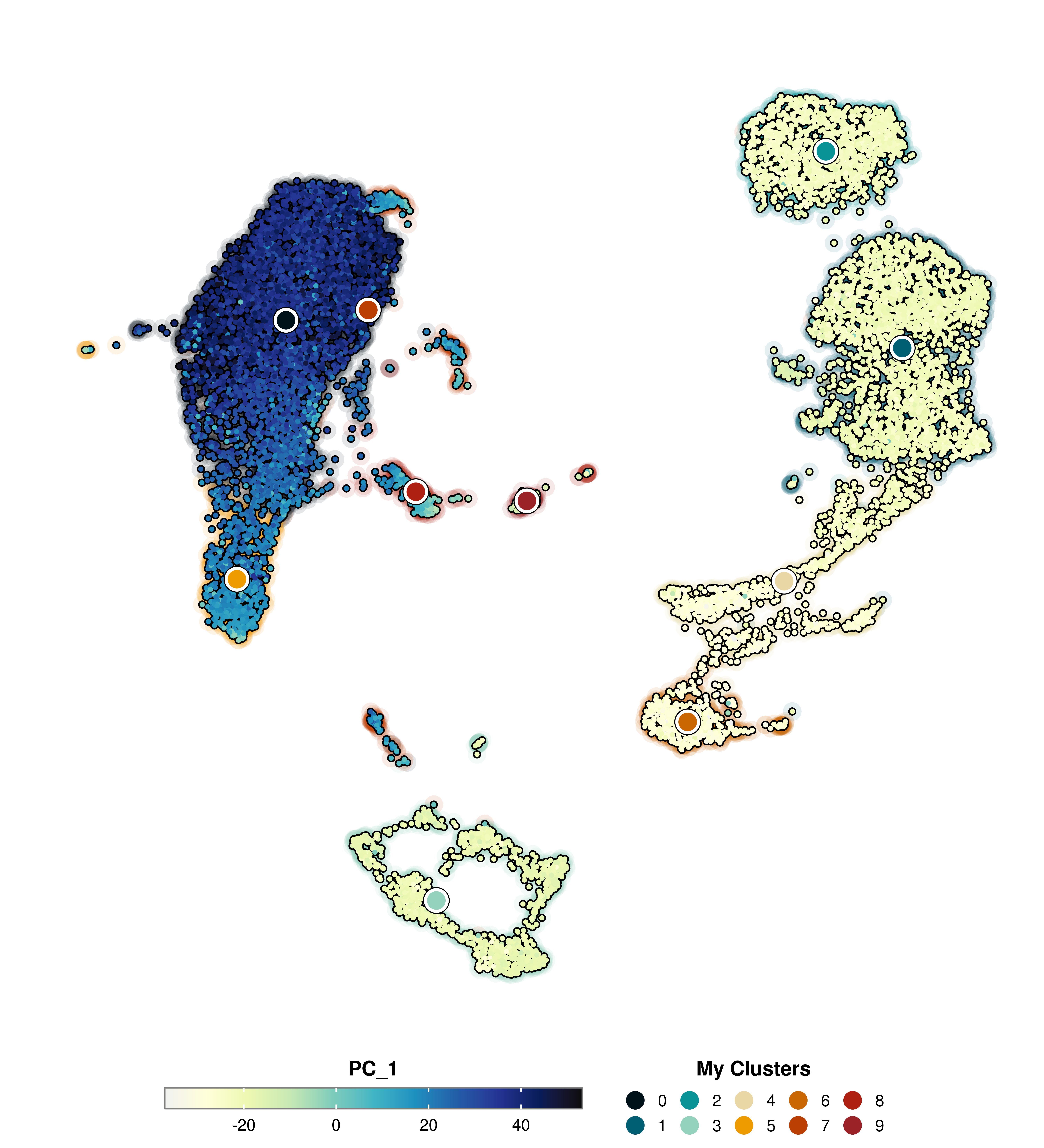
SCpubr::do_FeaturePlot(sample = sample,
features = "PC_1",
label = TRUE,
label.color = "#ca6702",
label.size = 8)